Services
Nucleic Acid automated Extraction
Human and other Organisms Genotyping

Nucleic Acid automated Extraction
Human and other Organisms Genotyping
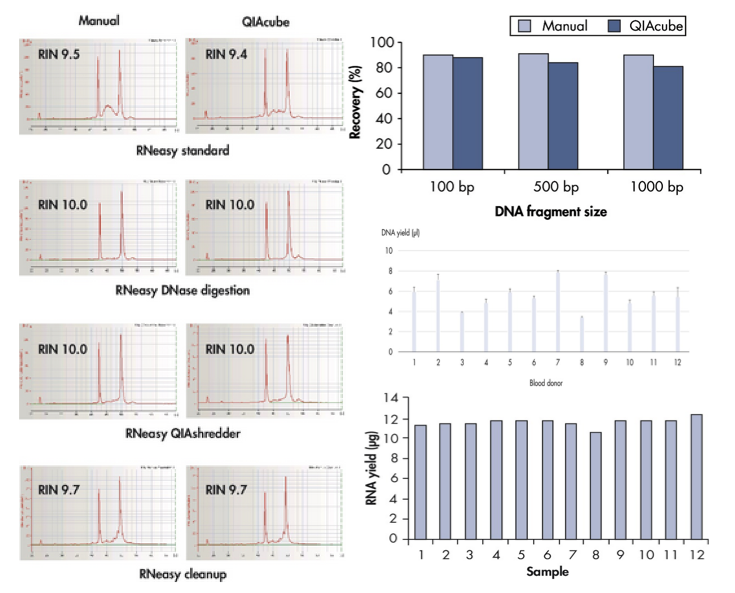
High-quality nucleic acids are a prerequisite for accurate and reliable genotyping and NGS results from complex samples. We offer the best solution for automated extraction of DNA and RNA, allowing you to extract nucleic acids in an unbiased manner without the loss of valuable genetic information.
QIAcube instrument is avaible in the Genotyping unit for extraction of DNA and RNA, offering you maximum confidence for processing precious samples for genotyping and sequencing. Automated processing of samples saves time, achieves standardization, reproducibility and consistency in your DNA and RNA extraction. Simultaneously, the sample preparation require minimal starting material, facilitate reduction of artifacts and increase the confidence and accuracy of your sequence data.
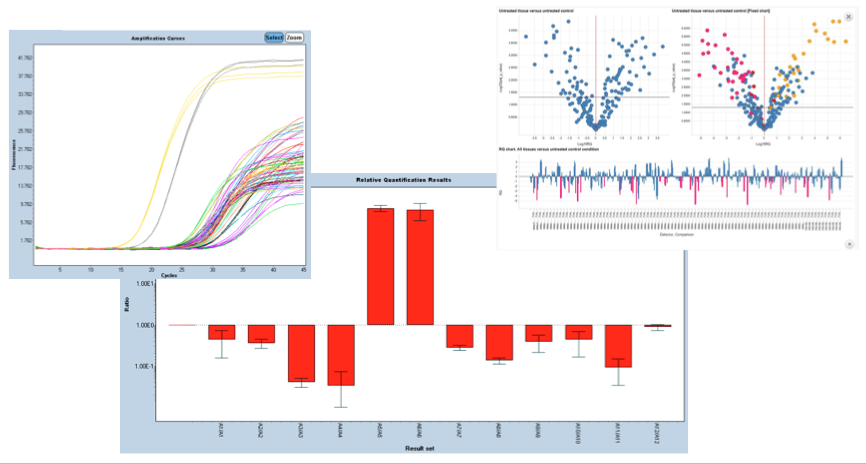
Gene expression assays are developed by relative quantification, which compares the levels of two different target sequences in a single sample, for instance a target gene of interest (GOI) and another gene (reference gene) and expresses the final result as a ratio of these targets.
Gene expression studies usually try to determine the way a target gene changes its expression profile over time (e.g., how much the expression changes in the course of a disease or treatment) relative to a defined starting point (e.g., disease-free or untreated state).
Additional Software: data analysis represents one of the biggest bottlenecks in qPCR experiments. For this reason, data statistical analysis is also avaible using Integromics´ RealTime StatMiner© software.
Viral load studies are developed by absolute quantification and allows scientists to quantify a single target sequence and express the final result as an absolute value (e.g., viral load - copies/ml). Such analyses is routinely used in areas like virology and microbiology.
 This unit offers a large variety of platforms to human and other organisms Genotyping and a customized platform could be employed according to user needs and depends of the objetive of the study.
This unit offers a large variety of platforms to human and other organisms Genotyping and a customized platform could be employed according to user needs and depends of the objetive of the study.
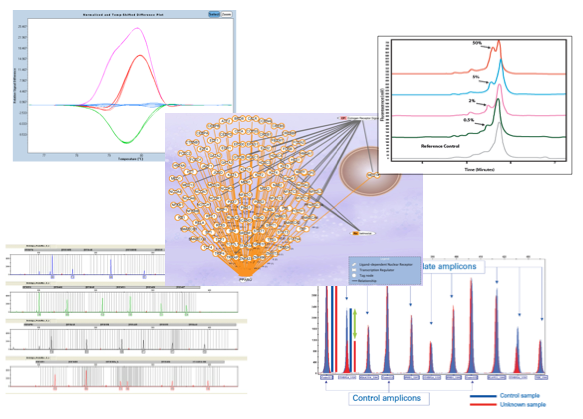
Single nucleotide polymorphisms (SNPs) and copy number polymorphisms (CNPs) assays are available using high-throughput detection systems. SNPs are the most common type of genetic variation in the human genome and could predispose people to disease (Human diagnosis) or influence their response to a drug (Pharmacogenetic). However, structural variation changes, such as deletion and duplications, referred to as CNPs are proving to be more common than expected. In animals and plants, SNPs also play an important role in the study of genetic variations within a population (Characterisation of genetic variability and Genetic improvement).
Addtional fragment analysis can be develop in our facilities. On the one hand, STRs (Short Tandem Repeats) or MPLAs (Multiplex Ligation-dependent Probe Amplification) assays, which can be used for Pedigree test and Detections of large delections or duplications in genomic DNA, respectively. On the other hand, AFLPs (Amplified Fragment Lengh Polymorphism) or MLST (Multi Locus Sequence Typing) assays are also avaible, and are widely used for Animal and plants identifications and Microbial typification, respectively.
Addtional software and Databases: depending of user needs, different softwares and databases such as Mutation Surveyor©, Alamut©, Genevestigator© or Ingenuity Pathways Analysis (IPA©) are offered to enhance the studies.
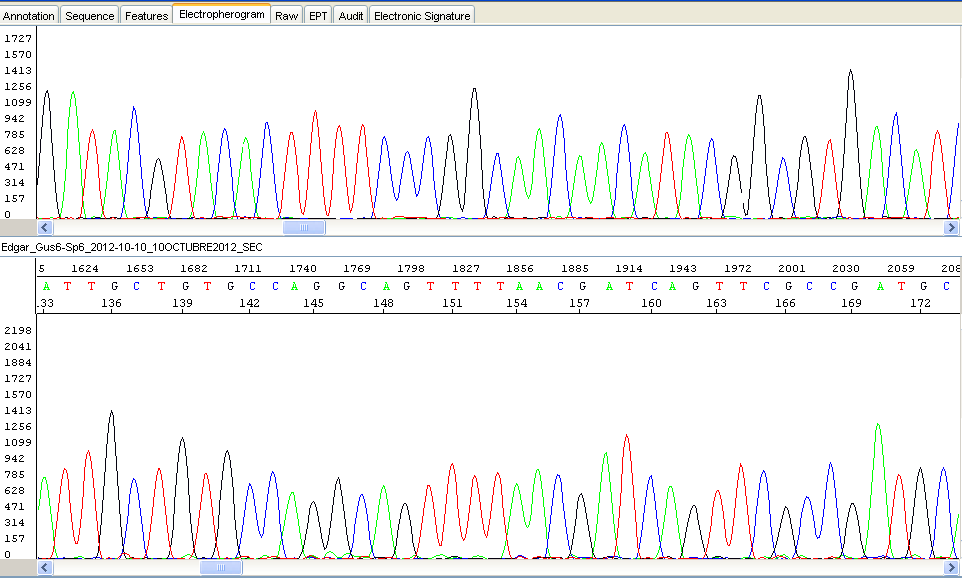
This unit offers a Sanger Sequencing Service. De novo sequencing and Resequencing are avaible. De novo sequencing is used for the generation of the DNA sequence of a DNA molecule without any prior information about the sequence. Resequencing is defined as sequencing of DNA molecules followed by comparison to a known or reference sequence, so resequencing is a method used to re-sequence targeted regions of a genome with the purpose of finding genetic variations or to proofread plasmid DNA sequence to verify the integrity of a gene inserted into an expression vector.
Sanger sequencing is most suitable for small to medium range sequencing projects. For large scale sequencing projects, please contact Ultrasequencing unit.